Sources
R packages
"ape" (Paradis et al. 2004)
"nlme" (Pinheiro et al. 2014)
"caper" (Orme et al. 2012)
"AICcmodavg" (Mazerolle 2013)
Data
Species-specific trait data ("primate_spec.txt"), a tab separated text file, species-specific data on brain size and body size in primates
Sample of phylogenetic trees ("10kTrees_Primates.nex"), 1000 phylogenetic hypotheses in nexus format, phylogeny of primates from Arnold et al. (2010)
A consensus tree ("consensusTre_Primates.nex"), the consenus phylogenetic tree for the above sample, nexus format, data from Arnold et al. (2010)
Codes
To get started
# activate libraries
library(ape)
library(nlme)
library(caper)
library(AICcmodavg)
# import data and trees
xdata <- read.table(file = "primate_spec.txt", header = T, sep = "\t", dec = ".")
row.names(xdata) = xdata$species
trees <- read.nexus("10kTrees_Primates.nex")
cons <- read.nexus("consensusTre_Primates.nex")
ape and nlme
We will run models on all trees then perform model averaging across them to get parameter estimates.
# for simplicity, we do not estimate lambda here
Cand.models = list()
niter = 1000
for (i in 1:niter) {
Cand.models[[i]] = gls(lg.brain ~ lg.body, data = xdata, method = "ML",
correlation = corPagel(1, trees[[i]], fixed = T))
}
# if you want to see the particular models
head(aictab(cand.set = Cand.models, modnames = paste("Tree Nr.", 1:niter, sep = " "),
sort = F), 20)
##
## Model selection based on AICc :
##
## K AICc Delta_AICc AICcWt LL
## Tree Nr. 1 3 57.33 6.47 0 -25.52
## Tree Nr. 2 3 55.53 4.67 0 -24.62
## Tree Nr. 3 3 56.37 5.51 0 -25.04
## Tree Nr. 4 3 57.56 6.70 0 -25.63
## Tree Nr. 5 3 56.26 5.40 0 -24.98
## Tree Nr. 6 3 55.09 4.23 0 -24.40
## Tree Nr. 7 3 55.49 4.63 0 -24.60
## Tree Nr. 8 3 57.04 6.18 0 -25.37
## Tree Nr. 9 3 56.38 5.52 0 -25.04
## Tree Nr. 10 3 57.52 6.66 0 -25.61
## Tree Nr. 11 3 56.73 5.87 0 -25.22
## Tree Nr. 12 3 53.81 2.95 0 -23.76
## Tree Nr. 13 3 56.19 5.34 0 -24.95
## Tree Nr. 14 3 57.02 6.16 0 -25.37
## Tree Nr. 15 3 57.36 6.50 0 -25.53
## Tree Nr. 16 3 55.63 4.77 0 -24.67
## Tree Nr. 17 3 57.23 6.37 0 -25.47
## Tree Nr. 18 3 56.49 5.63 0 -25.10
## Tree Nr. 19 3 58.61 7.75 0 -26.16
## Tree Nr. 20 3 55.40 4.54 0 -24.55
# model averaged intercept
Int_av = modavg(parm = "(Intercept)", cand.set = Cand.models, modnames = paste("Tree Nr.",
1:niter, sep = " "))
Int_av$Mod.avg.beta
# model averaged slope
beta_av = modavg(parm = "lg.body", cand.set = Cand.models, modnames = paste("Tree Nr.",
1:niter, sep = " "))
beta_av$Mod.avg.beta
caper
"AICcmodavg" does not work in this case, we need to do the multomodel inference "by hand". See also OPM for "1) Selecting among evolutionary models with different combinations of predictors"
# run all models, for simplicity we do not estimate lambda here
params = c()
niter = 1000
for (i in 1:niter) {
tree.data = comparative.data(phy = trees[[i]], data = xdata, names.col = species,
vcv = T, na.omit = F, warn.dropped = T)
mod = pgls(lg.brain ~ lg.body, data = tree.data, lambda = 1)
params = rbind(params, data.frame(row.names = NULL, tree = i, logLik = mod$model$log.lik,
AIC = mod$aicc, lambda = mod$param[2], Int = mod$model$coef[1], Int_err = mod$sterr[1],
beta = mod$model$coef[2], beta_err = mod$sterr[2]))
}
# get AIC delta and w
params$delta = params$AIC - min(params$AIC)
params$w = exp(-0.5 * params$delta)/sum(exp(-0.5 * params$delta))
# model averaging of parameters of interest
weighted.mean(params$Int, params$w) #averaged Intercept
w_err = c()
for (i in 1:niter) {
w_err[i] = sqrt(params[i, ]$Int_err^2 + (params[i, ]$Int - weighted.mean(params$Int,
params$w))^2)
}
weighted.mean(w_err, params$w) #SE
weighted.mean(params$Int, params$w) - 1.96 * weighted.mean(w_err, params$w) #lower CI (from SE)
weighted.mean(params$Int, params$w) + 1.96 * weighted.mean(w_err, params$w) #Upper CI (from SE)
quantile(params$Int, c(0.025, 0.975)) #CI
## 2.5% 97.5%
## 1.075 1.288
weighted.mean(params$beta, params$w) #averaged slope
for (i in 1:niter) {
w_err[i] = sqrt(params[i, ]$beta_err^2 + (params[i, ]$beta - weighted.mean(params$beta,
params$w))^2)
}
weighted.mean(w_err, params$w) #SE
weighted.mean(params$beta, params$w) - 1.96 * weighted.mean(w_err, params$w) #Lower CI
weighted.mean(params$beta, params$w) + 1.96 * weighted.mean(w_err, params$w) #Upper CI
quantile(params$beta, c(0.025, 0.975)) #CI
## 2.5% 97.5%
## 0.2778 0.3128
Plotting the results
We will plot all models corresponding to different trees
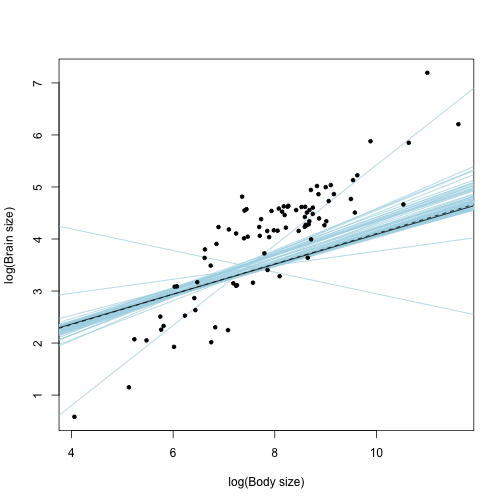
plot(xdata$lg.body, xdata$lg.brain, xlab = "log(Body size)", ylab = "log(Brain size)",
pch = NA_integer_)
# this draws the regression lines corresponding to different trees in the sample:
for (i in 1:niter) {
abline(Cand.models[[i]], col = "lightblue")
}
# this is what you would get if you used the consensus phylogenetic tree:
abline(gls(lg.brain ~ lg.body, data = xdata, method = "ML", correlation = corPagel(1,
cons, fixed = T)))
# this is what the model averaged parameters deliniate:
abline(a = Int_av$Mod.avg.beta, b = beta_av$Mod.avg.beta, lty = 2)
points(xdata$lg.body, xdata$lg.brain, pch = 20)
We can see that different trees define slightly different regression lines (light blue). However, with the exception of few exteremes, most of them vary along the same mean (that can be characterized by the model averaged -dashed- line or the -solid- line that corresponds to the consensus tree). This figure suggests only a small confounding role for phylogenetic uncertainty.
References
- Arnold C, Matthews LJ, Nunn CL (2010) The 10kTrees website: a new online resource for primate hylogeny. Evol Anthropol 19:114-118.
- Mazerolle MJ (2013) AICcmodavg: Model selection and multimodel inference based on (Q)AIC(c). R package version 1.28.
- Orme D, Freckleton R, Thomas G, Petzoldt T, Fritz S, Isaac N, Pearse W (2012) caper: Comparative Analyses of Phylogenetics and Evolution in R. R package version 3.1-104.
- Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289-290.
- Pinheiro J, Bates D, DebRoy S, Sarkar D, R Development Core Team (2012) nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-104,