Sources
R packages
"ape" (Paradis et al. 2004)
"nlme" (Pinheiro et al. 2014)
"AICcmodavg" (Mazerolle 2013)
"geiger" (Harmon et al. 2008)
Data
Species-specific trait data ("primate_spec.txt"), a tab separated text file, species-specific data on brain size and body size in primates, include within-species sample sizes
Phylogeny ("consensusTre_Primates.nex"), the consenus phylogenetic tree for 1000 phylogenetic hypotheses from Arnold et al. (2010), nexus format
Codes
To get started
# activate libraries
library(ape)
library(nlme)
library(AICcmodavg)
library(geiger)
# import data and tree
xdata <- read.table(file = "primate_spec.txt", header = T, sep = "\t", dec = ".")
row.names(xdata) = xdata$species
tree <- read.nexus("consensusTre_Primates.nex")
Creating different models considering different confounding effects
Ordinary least square (OLS) regression not considering phylogeny and data heterogeneity
# star phylogeny
tree0 <- rescale(tree, "lambda" , 0)
bm.tree0 <- corBrownian(phy = tree0)
# same weight for all species
xdata$constant = rep(1, length(xdata$brain_N))
# these are all equivalent models
ols = gls(lg.brain ~ lg.body, xdata, correlation = bm.tree0, weights = ~constant,
method = "ML")
ols.1 = gls(lg.brain ~ lg.body, xdata, method = "ML")
ols.2 = lm(lg.brain ~ lg.body, xdata)
Weighted OLS considering heterogeneity in data quality by using unequal weights
# these are all equivalent models
w.ols = gls(lg.brain ~ lg.body, xdata, correlation = bm.tree0, weights = ~1/brain_N,
method = "ML")
w.ols.1 = gls(lg.brain ~ lg.body, xdata, weights = ~1/brain_N, method = "ML")
w.ols.2 = lm(lg.brain ~ lg.body, xdata, weights = brain_N)
Phylogenetic OLS considering phylogenetic relationships of species
# we can consider different models for trait evolution
pgls.bm = gls(lg.brain ~ lg.body, xdata, correlation = corBrownian(phy = tree),
weights = ~constant, method = "ML")
pgls.pa = gls(lg.brain ~ lg.body, xdata, correlation = corPagel(1, phy = tree),
weights = ~constant, method = "ML")
Weighted PGLS considering both phylogeny and data heterogeneity
w.pgls.bm = gls(lg.brain ~ lg.body, xdata, correlation = corBrownian(phy = tree),
weights = ~1/brain_N, method = "ML")
w.pgls.pa = gls(lg.brain ~ lg.body, xdata, correlation = corPagel(1, phy = tree),
weights = ~1/brain_N, method = "ML")
Selecting the model that offers the best fit relative to model complexity
Models are only comparable if are fitted by Maximum Likelihood!
Cand.models = list()
Cand.models[[1]] = ols
Cand.models[[2]] = w.ols
Cand.models[[3]] = pgls.bm
Cand.models[[4]] = pgls.pa
Cand.models[[5]] = w.pgls.bm
Cand.models[[6]] = w.pgls.pa
Modnames = paste(c("ols", "w.ols", "pgls.bm", "pgls.pa", "w.pgls.bm", "w.pgls.pa"),
sep = " ")
aictab(cand.set = Cand.models, modnames = Modnames, sort = T)
##
## Model selection based on AICc :
##
## K AICc Delta_AICc AICcWt Cum.Wt LL
## pgls.pa 4 55.07 0.00 0.65 0.65 -23.29
## pgls.bm 3 56.33 1.26 0.35 1.00 -25.02
## ols 3 133.20 78.13 0.00 1.00 -63.45
## w.pgls.pa 4 158.35 103.29 0.00 1.00 -74.93
## w.ols 3 173.95 118.89 0.00 1.00 -83.83
## w.pgls.bm 3 176.66 121.60 0.00 1.00 -85.19
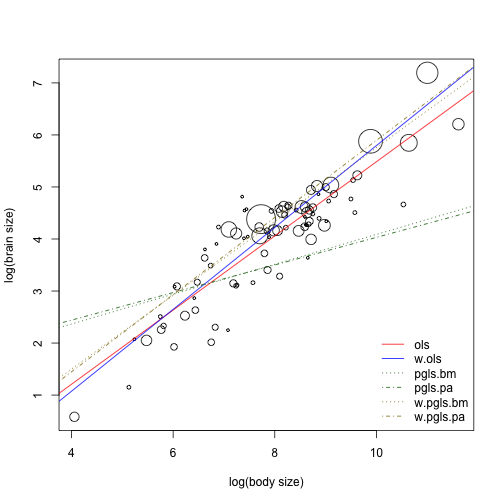
The best models are the PGLS models, weighting only worsens model fit (probably because of the high repeatability of traits).
Plotting the results
plot(xdata$lg.body, xdata$lg.brain, pch = 1, cex = 0.5 * sqrt(xdata$brain_N),
xlab = "log(body size)", ylab = "log(brain size)")
abline(ols, col = "red", lty = 1)
abline(w.ols, col = "blue", lty = 1)
abline(pgls.bm, col = "darkgreen", lty = 3)
abline(pgls.pa, col = "darkgreen", lty = 4)
abline(w.pgls.bm, col = "gold4", lty = 3)
abline(w.pgls.pa, col = "gold4", lty = 4)
legend(x = "bottomright", legend = c("ols", "w.ols", "pgls.bm", "pgls.pa", "w.pgls.bm",
"w.pgls.pa"), lty = c(1, 1, 3, 4, 3, 4), col = c("red", "blue", "darkgreen",
"darkgreen", "gold4", "gold4"), bty = "n")
The "weights of weights": the "omega" scaling factor
In the above models, we used the untransformed species-specific (within-species) sample sizes as statistical weights. However, as we argued in our chapter, this option may irrealistically overemphasize the influence of intensively studied species with large sample sizes. To downweight such unwanted effects one can apply transformations on the species-specific sample sizes. In chapter 12, we introduced the parameter "omega" that defines the function for such transformation. Omega is simply an elevation factor that ranges from 1 to 1/100 and characterises the exponent of the within-species sample sizes (i.e. if omega is 1, the original sample sizes are used as weights, if it is 1/2=0.5, the square-root-transformed values serve as weights, if omega is 1/100 it approximates to 0 thus represents the scenario in which all species are considered with equal weight (n^0 = 1). In the example below, we demonstrate how one can obtain parameter estimates by averaging across different models relying on different omegas by considering their fit to the data.
# all models from omega=1 to omega=0
Cand.models.w = list()
xweights = seq(1, 0, length.out = 101)
for (w in 1:length(xweights)) {
omega = xdata$brain_N^xweights[w]
Cand.models.w[[w]] = gls(lg.brain ~ lg.body, data = xdata, correlation = corPagel(1,
phy = tree), weights = ~1/omega, method = "ML")
}
modnames.w = paste("omega =", round(xweights, 2), sep = " ")
# model averaged inetercept
Int_av = modavg(parm = "(Intercept)", cand.set = Cand.models.w, modnames = modnames.w)
Int_av$Mod.avg.beta
# model averaged slope
beta_av = modavg(parm = "lg.body", cand.set = Cand.models.w, modnames = modnames.w)
beta_av$Mod.avg.beta
# if you want to see the particular models
head(aictab(cand.set = Cand.models.w, modnames = modnames.w, sort = F), 100)
##
## Model selection based on AICc :
##
## K AICc Delta_AICc AICcWt LL
## omega = 1 4 158.35 103.29 0.00 -74.93
## omega = 0.99 4 157.23 102.16 0.00 -74.37
## omega = 0.98 4 156.11 101.04 0.00 -73.81
## omega = 0.97 4 155.00 99.93 0.00 -73.25
## omega = 0.96 4 153.89 98.82 0.00 -72.70
## omega = 0.95 4 152.79 97.72 0.00 -72.15
## omega = 0.94 4 151.69 96.63 0.00 -71.60
## omega = 0.93 4 150.60 95.54 0.00 -71.05
## omega = 0.92 4 149.52 94.45 0.00 -70.51
## omega = 0.91 4 148.44 93.37 0.00 -69.97
## omega = 0.9 4 147.36 92.30 0.00 -69.44
## omega = 0.89 4 146.30 91.23 0.00 -68.90
## omega = 0.88 4 145.24 90.17 0.00 -68.37
## omega = 0.87 4 144.18 89.11 0.00 -67.84
## omega = 0.86 4 143.13 88.06 0.00 -67.32
## omega = 0.85 4 142.09 87.02 0.00 -66.80
## omega = 0.84 4 141.05 85.98 0.00 -66.28
## omega = 0.83 4 140.01 84.95 0.00 -65.76
## omega = 0.82 4 138.99 83.92 0.00 -65.25
## omega = 0.81 4 137.97 82.90 0.00 -64.74
## omega = 0.8 4 136.95 81.89 0.00 -64.23
## omega = 0.79 4 135.94 80.88 0.00 -63.72
## omega = 0.78 4 134.94 79.87 0.00 -63.22
## omega = 0.77 4 133.94 78.87 0.00 -62.72
## omega = 0.76 4 132.95 77.88 0.00 -62.23
## omega = 0.75 4 131.96 76.89 0.00 -61.73
## omega = 0.74 4 130.98 75.91 0.00 -61.24
## omega = 0.73 4 130.00 74.94 0.00 -60.75
## omega = 0.72 4 129.03 73.96 0.00 -60.27
## omega = 0.71 4 128.06 73.00 0.00 -59.78
## omega = 0.7 4 127.10 72.04 0.00 -59.30
## omega = 0.69 4 126.15 71.08 0.00 -58.83
## omega = 0.68 4 125.20 70.13 0.00 -58.35
## omega = 0.67 4 124.25 69.18 0.00 -57.88
## omega = 0.66 4 123.31 68.24 0.00 -57.41
## omega = 0.65 4 122.37 67.30 0.00 -56.94
## omega = 0.64 4 121.43 66.37 0.00 -56.47
## omega = 0.63 4 120.50 65.44 0.00 -56.01
## omega = 0.62 4 119.58 64.51 0.00 -55.54
## omega = 0.61 4 118.66 63.59 0.00 -55.08
## omega = 0.6 4 117.73 62.67 0.00 -54.62
## omega = 0.59 4 116.82 61.75 0.00 -54.16
## omega = 0.58 4 115.90 60.84 0.00 -53.70
## omega = 0.57 4 114.99 59.92 0.00 -53.25
## omega = 0.56 4 114.08 59.01 0.00 -52.79
## omega = 0.55 4 113.17 58.10 0.00 -52.34
## omega = 0.54 4 112.25 57.19 0.00 -51.88
## omega = 0.53 4 111.34 56.28 0.00 -51.42
## omega = 0.52 4 110.43 55.36 0.00 -50.97
## omega = 0.51 4 109.52 54.45 0.00 -50.51
## omega = 0.5 4 108.60 53.53 0.00 -50.05
## omega = 0.49 4 107.68 52.61 0.00 -49.59
## omega = 0.48 4 106.75 51.69 0.00 -49.13
## omega = 0.47 4 105.82 50.76 0.00 -48.66
## omega = 0.46 4 104.88 49.82 0.00 -48.19
## omega = 0.45 4 103.94 48.87 0.00 -47.72
## omega = 0.44 4 102.98 47.92 0.00 -47.24
## omega = 0.43 4 102.02 46.95 0.00 -46.76
## omega = 0.42 4 101.05 45.98 0.00 -46.28
## omega = 0.41 4 100.06 44.99 0.00 -45.78
## omega = 0.4 4 99.06 44.00 0.00 -45.28
## omega = 0.39 4 98.06 42.99 0.00 -44.78
## omega = 0.38 4 97.04 41.97 0.00 -44.27
## omega = 0.37 4 96.00 40.94 0.00 -43.75
## omega = 0.36 4 94.96 39.89 0.00 -43.23
## omega = 0.35 4 93.90 38.84 0.00 -42.71
## omega = 0.34 4 92.84 37.77 0.00 -42.17
## omega = 0.33 4 91.76 36.69 0.00 -41.63
## omega = 0.32 4 90.67 35.60 0.00 -41.09
## omega = 0.31 4 89.57 34.50 0.00 -40.54
## omega = 0.3 4 88.45 33.39 0.00 -39.98
## omega = 0.29 4 87.33 32.26 0.00 -39.42
## omega = 0.28 4 86.19 31.12 0.00 -38.85
## omega = 0.27 4 85.04 29.97 0.00 -38.27
## omega = 0.26 4 83.88 28.82 0.00 -37.69
## omega = 0.25 4 82.71 27.64 0.00 -37.11
## omega = 0.24 4 81.53 26.46 0.00 -36.52
## omega = 0.23 4 80.33 25.27 0.00 -35.92
## omega = 0.22 4 79.13 24.06 0.00 -35.32
## omega = 0.21 4 77.91 22.84 0.00 -34.71
## omega = 0.2 4 76.68 21.62 0.00 -34.09
## omega = 0.19 4 75.44 20.38 0.00 -33.48
## omega = 0.18 4 74.20 19.13 0.00 -32.85
## omega = 0.17 4 72.94 17.87 0.00 -32.22
## omega = 0.16 4 71.67 16.60 0.00 -31.59
## omega = 0.15 4 70.40 15.33 0.00 -30.95
## omega = 0.14 4 69.12 14.05 0.00 -30.31
## omega = 0.13 4 67.83 12.77 0.00 -29.67
## omega = 0.12 4 66.55 11.48 0.00 -29.03
## omega = 0.11 4 65.27 10.21 0.00 -28.39
## omega = 0.1 4 64.01 8.94 0.00 -27.76
## omega = 0.09 4 62.76 7.70 0.01 -27.13
## omega = 0.08 4 61.55 6.49 0.01 -26.53
## omega = 0.07 4 60.39 5.33 0.02 -25.95
## omega = 0.06 4 59.30 4.23 0.03 -25.40
## omega = 0.05 4 58.30 3.23 0.05 -24.90
## omega = 0.04 4 57.41 2.34 0.08 -24.46
## omega = 0.03 4 56.65 1.58 0.12 -24.08
## omega = 0.02 4 56.02 0.95 0.17 -23.76
## omega = 0.01 4 55.50 0.44 0.22 -23.51
From this, it is also clear that in the current case, weighting only worsens model fit (probably because of the high repeatability of traits).
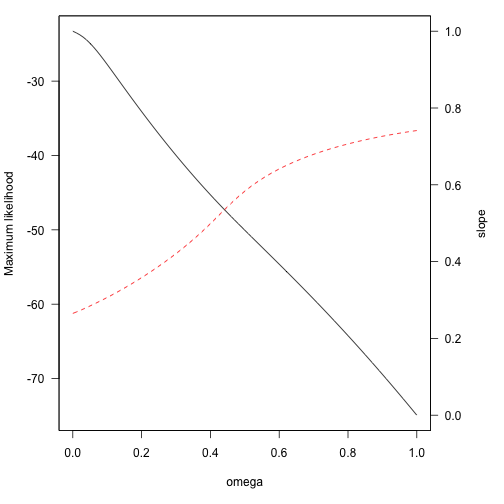
Let us make a figure to summarize model fit and parameter estimates at different omegas.
ll = unlist(lapply(Cand.models.w, function(x) {
x$logLik[1]
}))
par(mar = c(5, 4, 1, 5) + 0.1)
plot(xweights, ll, type = "l", xlab = "omega", ylab = "Maximum likelihood",
las = 1)
par(new = TRUE)
plot(xweights, unlist(lapply(Cand.models.w, coefficients))[seq(2, 202, 2)],
type = "l", col = "red", lty = 2, xaxt = "n", yaxt = "n", xlab = "", ylab = "",
ylim = c(0, 1))
axis(4, las = 1)
mtext("slope", side = 4, line = 3)
The highest Maximum Likelihood (solid black line) corresponds to omega = 0, which considers all species with equal weight and gives a slope estimate of 0.27. Different transformations on the within-species sample sizes (different omegas) would provide considerably different -but highly unrealistic- slope estimates (dashed red line).
References
- Arnold C, Matthews LJ, Nunn CL (2010) The 10kTrees website: a new online resource for primate hylogeny. Evol Anthropol 19:114-118.
- Harmon LJ, Weir J, Brock C, Glor RE, Challenger W (2008) GEIGER: Investigating evolutionary radiations. Bioinformatics 24:129-131.
- Mazerolle MJ (2013) AICcmodavg: Model selection and multimodel inference based on (Q)AIC(c). R package version 1.28.
- Orme D, Freckleton R, Thomas G, Petzoldt T, Fritz S, Isaac N, Pearse W (2012) caper: Comparative Analyses of Phylogenetics and Evolution in R. R package version 3.1-104.
- Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289-290.
- Pinheiro J, Bates D, DebRoy S, Sarkar D, R Development Core Team (2012) nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-104,