Sources
R packages
"ape" (Paradis et al. 2004)
"car" (Fox & Weisberg 2011)
"caper" (Orme et al. 2013)
Data
Phylogenic tree ("fict_phylo.nex"), Nexus file
Trait data ("covariates.txt"), tab-separated txt-file
Functions
Functions used in the practical ("pgls_check_fncs.r"), r file
Codes
Codes
The data set used as an example throughout is fictitious to avoid confusion with real data (and to have all the properties desired for the purpose of this practical). It consists of a phylogeny comprising 100 taxa. The phylogeny is a real one, in the nexus format, and taken from the primate tree (version 3) of the 10kTrees website (http://10ktrees.fas.harvard.edu/; Arnold et al. 2010). The names are altered, though, to avoid confusion with real data. The traits are fictitious, too.
The model aims at investigating the effects of group size, home range size, longevity and mating system on brain size. Body size should be controlled for because of the allometric relation between body size and brain size.
Getting started with the phylogenetic tree:
#load library ape:
library(ape)
#read the phylogeny into an R object (you will need to set the appropriate working directory first using setwd()):
fict.phylo=read.nexus("fict_phylo.nex")
#inspect the resulting object:
str(fict.phylo)
## List of 4
## $ edge : int [1:197, 1:2] 101 102 103 104 105 106 107 108 109 110 ...
## $ edge.length: num [1:197] 4.16 22.03 16.81 8.59 6.54 ...
## $ Nnode : int 98
## $ tip.label : chr [1:100] "Sucehtipocrec_siralugobla" "Sucehtipocrec_suhpec" "Sucehtipocrec_itseohl" "Sucehtipocrec_anom" ...
## - attr(*, "class")= chr "phylo"
## - attr(*, "order")= chr "cladewise"
#>= 'fict.phylo' is a tree with 100 tips and 98 internal nodes
#plot the tree:
plot.phylo(x=fict.phylo, cex=0.7, direction="upwards", no.margin=TRUE)
Getting started with the traits data:
#read data with characters of the fictitious species:
covariates=read.table(file="covariates.txt", header=T, sep="\t")
#inspect the resulting object:
str(covariates)
## 'data.frame': 100 obs. of 7 variables:
## $ species : Factor w/ 100 levels "Acacam_anaeknot",..: 70 71 69 66 67 68 59 58 72 51 ...
## $ group.size : num 9.4 9 5.8 7.8 3.4 3.9 5.8 5.7 2.5 26.9 ...
## $ hr.size : num 37.1 32.8 31.5 29.7 32.2 ...
## $ body.size : num 1.89 1.13 1.11 0.96 1.02 1.06 2.25 1.4 0.87 5.6 ...
## $ longevity : num 38.5 37.9 40.3 43.3 43.4 ...
## $ mating.system: Factor w/ 4 levels "multiF.multiM",..: 4 2 4 2 2 3 4 1 2 1 ...
## $ brain.size : num 14.21 11.52 8.62 8.17 7.24 ...
#=> 'covariates' is a data frame with 100 observations (rows) and 7 variables (columns)
#four of the variables are quantitative predictors, namely, 'group.size', 'hr.size' (home range size),
#'body.size', and 'longevity', one ('mating.system') is a factor (qualitative or categorical predictor)
#with four levels
#the response is 'brain.size'
Inspect and prepare the data frame with the traits:
#check whether there are any missing values in 'covariates':
sum(is.na(covariates))
#=> the data set is complete (if it were not, one should keep only those rows that are free of missing values
#in the variables needed)
#inspect the distributions of the quantitative predictor variables:
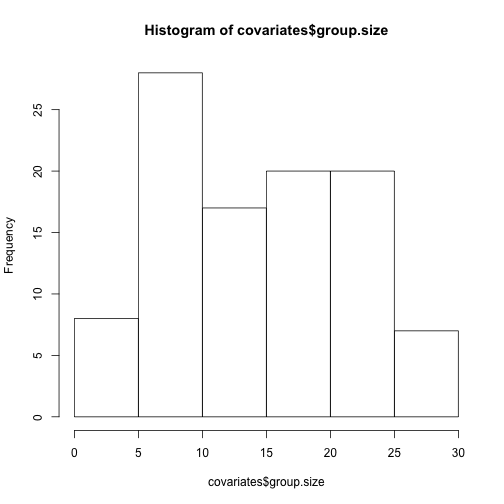
hist(covariates$group.size)#looks okay (not heavily skewed, no outliers)
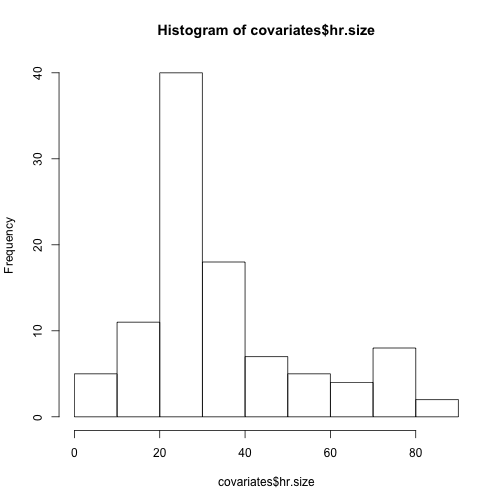
hist(covariates$hr.size)#looks okay (not heavily skewed, no outliers)
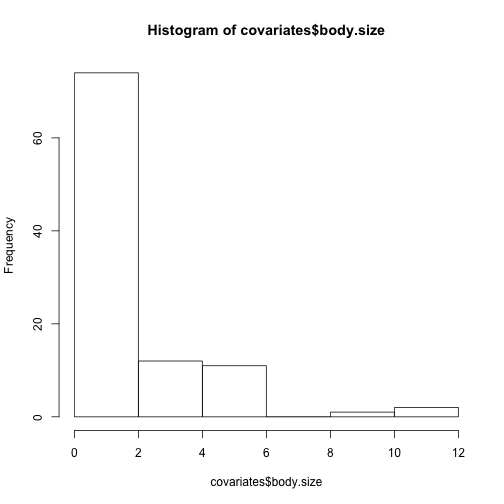
hist(covariates$body.size)#is pretty skewed;
#to avoid influential cases check whether 'body.size' looks more symmetrical after a log-transformation:
#first check whether the minimum is >0 (which it should be for a variable like body size):
min(covariates$body.size)#it is
hist(log(covariates$body.size))#looks okay
#get log-transformed body size into the data frame 'covariates':
covariates$log.body.size=log(covariates$body.size)
hist(covariates$longevity)#looks okay (not heavily skewed, no outliers)
#inspect the frequency distribution of the factor 'mating.system':
table(covariates$mating.system)
##
## multiF.multiM multiF.uniM multiM.uniF uniM.uniF
## 30 33 2 35
#one level ('multiM.uniF') is represented by only two species; this is somewhat problematic because the two species
#are likely to be particularly influential and at the same time probably not a very good representation of this
#mating system
#=> omit them from the data:
sel.covariates=subset(x=covariates, mating.system!="multiM.uniF")
#check whether this was effective:
nrow(sel.covariates)#reveals 98; two rows have been dropped
#frequently it is a good idea to get rid of levels of factors not present in the data any more;
#to achieve this use:
sel.covariates=droplevels(x=sel.covariates)
#after having omitted species from 'covariates' one needs to also drop them from the phylogenetic tree
#before doing so one should check whether the names of the taxa included in the tree and the data are spelled
#identically:
length(levels(covariates$species))#number of different species in 'covariates' is 100
length(fict.phylo$tip.label)#number of species in 'fict.phylo' is 100
length(intersect(levels(covariates$species), fict.phylo$tip.label))
#the intersection of the two is also 100 (typos, missing species, etc. would lead to a smaller number)
#now drop those species from the tree which are no more in the data:
sel.tree=drop.tip(phy=fict.phylo, tip=fict.phylo$tip.label[!fict.phylo$tip.label%in%as.character(sel.covariates$species)], rooted=T)
#inspect the resulting tree:
str(sel.tree)#96 nodes are left
## List of 4
## $ edge : int [1:193, 1:2] 99 100 101 102 103 104 105 106 107 108 ...
## $ edge.length: num [1:193] 4.16 22.03 16.81 8.59 6.54 ...
## $ Nnode : int 96
## $ tip.label : chr [1:98] "Sucehtipocrec_siralugobla" "Sucehtipocrec_suhpec" "Sucehtipocrec_itseohl" "Sucehtipocrec_anom" ...
## - attr(*, "class")= chr "phylo"
## - attr(*, "order")= chr "cladewise"
#finally, inspect the distribution of the response:
hist(sel.covariates$brain.size)#looks okay (not heavily skewed, no outliers)
Dealing with collinearity:
lm.res=lm(brain.size ~ group.size + hr.size + longevity + log.body.size + mating.system, data=sel.covariates)
library(car)
## Loading required package: MASS
## Loading required package: nnet
## GVIF Df GVIF^(1/(2*Df))
## group.size 2.576 1 1.605
## hr.size 11.963 1 3.459
## longevity 2.598 1 1.612
## log.body.size 11.640 1 3.412
## mating.system 1.055 2 1.013
#=> reveals two pretty large Generalized Variance Inflation Factors (column GVIF) for home range and body size
#(a value of 10 or larger is commonly considered indicative of a problem)
#from the initial model I would drop home range size since brain size arguably is the more important predictor
#(but later I'd check the results of two additional models, one including both predictors in addition to all others,
#and one in which home range size substitutes body size)
#check whether the collinearity issue is removed when longevity is removed from the model:
lm.res=lm(brain.size ~ group.size + longevity + log.body.size + mating.system, data=sel.covariates)
vif(mod=lm.res)#it is
## GVIF Df GVIF^(1/(2*Df))
## group.size 2.488 1 1.577
## longevity 2.371 1 1.540
## log.body.size 1.406 1 1.186
## mating.system 1.019 2 1.005
Fitting the PGLS model using the function pgls of the package caper (Orme et al. 2013):
#the function pgls needs the data and the tree to be comprised in a single object which can be achieved using
#the function comparative.data of the package caper:
library(caper)
## Loading required package: mvtnorm
test.data=comparative.data(phy=sel.tree, data=sel.covariates, names.col=species, vcv=TRUE, na.omit=FALSE, warn.dropped=TRUE)
#now the PGLS model can be fitted:
full=pgls(brain.size ~ group.size + longevity + log.body.size + mating.system, data=test.data, lambda="ML")
#note that the argument lambda="ML" means that lambda, the scaling parameter of the variance-covariance matrix,
#is estimated (using Maximum Likelihood; one could also set it to a fixed value, e.g., 1 or 0)
Checking assumptions about the residuals:
#inspect phylogenetic residuals for normality:
#option 1, inspect a histogram:
hist(full$phyres)#looks pretty good (i.e., pretty normally distributed)
#option 2, quantile-quantile (QQ) plot (allows easier assessment of whether the assumption of normally distributed
#residuals is fulfilled):
qqnorm(full$phyres)
qqline(full$phyres)#looks pretty good
#note that if the assumption is fulfilled all points fall close to the line
#inspect phylogenetic residuals for homogeneity:
plot(x=fitted(full), y=full$phyres, pch=19)#looks pretty good
#note that in this latter plot no pattern whatsoever should be discernible
#it is also possible to get the three diagnostics plots at once using the function diagnostics.plot provided
#in the file pgls_check_fncs.r
#this requires to first 'source' the file (to make the functions in it available in R):
source("pgls_check_fncs.r")#(you will need to specify the right 'working directory')
## Loading required package: geiger
#then the diagnostics plots can be got using:
diagnostics.plot(full)
Absence of Influential cases is evaluated by excluding taxa, one at a time, from the data, rerunning the model and comparing the estimates derived with those obtained for the full data set. For convenience I programmed a function doing this. Its sole argument is a PGLS-model result obtained using the function pgls of the package caper. It returns a named list comprising the following objects:
summary: matrix with one row for each estimated model coefficient and one column for each of the estimate obtained for the full data set as well as the minimum and maximum of the estimates revealed after the case-wise deletion of taxa.
taxa.failed: single integer indicating the number of taxa for which the function pgls failed to converge when it was excluded.
estimates: matrix with one row for each taxon and one column for each of the estimates obtained after the of the respective taxon. This matrix one will need to inspect in more detail in case the model appeared to be unstable (as indicated by the output in 'summary') and one wants to identify the taxa responsible for this.
SEs: the same as 'estimates' but depicting the standard errors (in the context of 'normal' linear models the stability of standard errors is usually not considered; but for those interested I provide them).
params: the same as 'estimates' but for the scaling parameters kappa, lambda, and delta. Note that all the three are always included although at most only one of them was estimated as part of the pgls.
#to use the function I provide, the file 'pgls_check_fncs.r' needs to be 'sourced':
source("pgls_check_fncs.r")#(you will need to specify the right 'working directory')
#call the function with the object that resulted from the call of the function pgls (note that this might take
#quite some time depending on the size of the data set; you might need to be patient and must not get worried
#if 'nothing happens'; in case of the model considered here it takes some 3 minutes on my notebook);
stability.ests=influence.pgls(pgls.res=full)
#inspect the results:
#begin with a check for how many taxa the function pgls failed to converge:
stability.ests$taxa.failed#for no taxon the function pgls failed to converge
#probably the summary is the most interesting then:
round(stability.ests$summary, digits=3)
## orig min max
## (Intercept) 6.414 5.983 6.820
## group.size 0.600 0.589 0.622
## longevity -0.028 -0.039 -0.016
## log.body.size 5.051 4.951 5.137
## mating.systemmultiF.uniM -0.061 -0.135 0.019
## mating.systemuniM.uniF 0.025 -0.051 0.096
#looks good with the exception of the two estimates obtained for mating system (which show even uncertainty
#regarding their sign; though they are pretty close to zero anyways; but compare with those obtained for longevity)
#note that the function round only serves in getting convenient looking output, it can and occasionally has to be
#dropped or called with another number of digits
#also inspect stability for the scaling parameter:
round(apply(X=stability.ests$params, MARGIN=2, FUN=range), digits=3)# the model is very stable with regard to it
## kappa lambda delta
## [1,] 1 1 1
## [2,] 1 1 1
#(note that only one of them, namely lambda, was actually estimated here)
#the respective estimate for the full data set can be obtained from the respective model output, here
full$param
## kappa lambda delta
## 1 1 1
#in case the individual estimates and standard errors are of interest use (potentially without rounding)
round(stability.ests$estimates, digits=3)#shows the estimates obtained for each individual taxon exluded
## (Intercept) group.size longevity
## Sucehtipocrec_siralugobla 6.444 0.598 -0.028
## Sucehtipocrec_suhpec 6.432 0.595 -0.026
## Sucehtipocrec_itseohl 6.450 0.600 -0.030
## Sucehtipocrec_anom 6.274 0.602 -0.025
## Sucehtipocrec_atsiruatep 6.414 0.601 -0.029
## Subecorolhc_surhtyregyp 6.401 0.600 -0.028
## Subecorolhc_sulatnat 6.509 0.600 -0.030
## Sucehtipoim_niopalat 6.445 0.595 -0.026
## Subecolla_sitohcirt 6.435 0.599 -0.029
## Rumeloeahcra_irojam 6.398 0.601 -0.028
## Ihava_reginal 6.384 0.601 -0.028
## Ihava_silatnedicco 6.395 0.600 -0.029
## Suelagoriehc_iyelssorc 6.420 0.598 -0.027
## Suelagoriehc_rojam 6.419 0.599 -0.028
## Suelagoriehc_suidem 6.442 0.597 -0.027
## Ainotnebuad_sisneiracsagadam 6.104 0.598 -0.025
## Rumelue_retnevirbur 6.427 0.600 -0.030
## Rumelapah_sumis 6.275 0.601 -0.023
## Irdni_irdni 6.441 0.598 -0.028
## Rumelipel_silceea 6.414 0.600 -0.028
## Rumelipel_suniletsum 6.707 0.591 -0.035
## Rumelipel_ilosanairdnar 6.633 0.592 -0.029
## Rumelipel_sutaduacifur 6.334 0.604 -0.028
## Rumelipel_silanoirtnetpes 5.983 0.605 -0.016
## Subecorcim_eahtreb 6.482 0.599 -0.030
## Subecorcim_sisnevalognob 6.380 0.601 -0.028
## Subecorcim_sufuroesirg 6.555 0.594 -0.029
## Subecorcim_eaylloj 6.396 0.599 -0.027
## Subecorcim_sunixoym 6.390 0.604 -0.030
## Subecorcim_isnommis 6.385 0.602 -0.029
## Renahp_reficruf 6.365 0.604 -0.028
## Sucehtiporp_ilereuqoc 6.600 0.595 -0.032
## Sucehtiporp_illasrettat 6.539 0.597 -0.031
## Sucehtiporp_ixuaerrev 6.698 0.594 -0.034
## Aicerav_arbur 6.253 0.607 -0.029
## Attauola_ayarac 6.432 0.599 -0.028
## Attauola_ataillap 6.416 0.600 -0.028
## Seleta_specicsuf 6.560 0.597 -0.031
## Seleta_sucsinap 6.671 0.592 -0.032
## Seletyhcarb_sedionhcara 6.518 0.599 -0.033
## Xirhtogal_ahcirtogal 6.261 0.604 -0.024
## Sutoa_sutalufni 6.573 0.604 -0.038
## Sutoa_specirgin 6.617 0.602 -0.037
## Xirhtillac_eailime 6.125 0.606 -0.022
## Xirhtillac_atallicinep 6.504 0.598 -0.031
## Xirhtillac_aeamgyp 6.370 0.600 -0.026
## Subec_snorfibla 6.510 0.595 -0.029
## Suniugas_xatsym 6.668 0.592 -0.033
## Suniugas_supideo 6.234 0.607 -0.026
## Suniugas_sutitrapirt 6.599 0.596 -0.032
## Irimias_sisneivilob 6.323 0.603 -0.027
## Sirol_sunairekkedyl 6.296 0.602 -0.025
## Sirol_sudargidrat 6.248 0.602 -0.024
## Subecitcyn_sisnelagneb 6.461 0.599 -0.030
## Subecitcyn_gnacuoc 6.325 0.602 -0.026
## Subecitcyn_sucinavaj 6.365 0.599 -0.026
## Subecitcyn_sisneganem 6.422 0.602 -0.028
## Omoh_sneipas 6.172 0.599 -0.018
## Setabolyh_irelleum 6.473 0.598 -0.029
## Sucsamon_ealleirbag 6.466 0.598 -0.029
## Sucsamon_sutusan 6.314 0.604 -0.028
## Sucsamon_ikis 6.480 0.600 -0.031
## Ognop_sueamgyp 6.402 0.600 -0.028
## Aicehtip_aicehtip 6.443 0.600 -0.029
## Acacam_sediotcra 6.374 0.601 -0.028
## Acacam_sipolcyc 6.411 0.600 -0.027
## Acacam_siralucicsaf 6.407 0.600 -0.028
## Acacam_ikceh 6.403 0.599 -0.028
## Acacam_attalum 6.362 0.598 -0.025
## Acacam_anirtsemen 6.425 0.600 -0.029
## Acacam_snecsergin 6.542 0.597 -0.032
## Acacam_sunelis 6.490 0.600 -0.032
## Acacam_sunavlys 6.413 0.599 -0.028
## Acacam_anaeknot 6.444 0.598 -0.028
## Oipap_sibuna 6.263 0.604 -0.026
## Oipap_sulahpeconyc 6.400 0.600 -0.028
## Oipap_sayrdamah 6.302 0.604 -0.028
## Oipap_sunisru 6.412 0.600 -0.028
## Subecewgnur_ijnupik 6.526 0.598 -0.031
## Suboloc_sisnelogna 6.464 0.599 -0.030
## Suboloc_susorellev 6.364 0.601 -0.027
## Subolocoilip_muronodrog 6.375 0.601 -0.028
## Subolocoilip_issuerp 5.989 0.622 -0.027
## Subolocoilip_sutartimofur 6.612 0.597 -0.035
## Subolocorp_surev 6.820 0.594 -0.039
## Xirhtagyp_aerenic 6.481 0.599 -0.030
## Sucehtiponihr_sulucnuva 6.270 0.608 -0.026
## Sucehtiponihr_ihcilerb 6.312 0.607 -0.028
## Sucehtipyhcart_iruocaled 6.372 0.599 -0.026
## Sucehtipyhcart_isiocnarf 6.270 0.589 -0.016
## Sucehtipyhcart_mutoal 6.443 0.599 -0.028
## Sucehtipyhcart_ieryahp 6.102 0.602 -0.019
## Sucitoue_sulutnagele 6.541 0.598 -0.031
## Ogalag_ieihcstam 6.472 0.599 -0.030
## Ogalag_sisnelagenes 6.351 0.603 -0.029
## Rumeloto_sutaduacissarc 6.383 0.600 -0.027
## Rumeloto_iittenrag 6.418 0.600 -0.028
## Suisrat_sunacnab 6.400 0.600 -0.028
## log.body.size mating.systemmultiF.uniM
## Sucehtipocrec_siralugobla 5.016 -0.057
## Sucehtipocrec_suhpec 5.048 -0.064
## Sucehtipocrec_itseohl 5.055 -0.062
## Sucehtipocrec_anom 5.043 -0.056
## Sucehtipocrec_atsiruatep 5.052 -0.062
## Subecorolhc_surhtyregyp 5.037 -0.069
## Subecorolhc_sulatnat 5.002 -0.114
## Sucehtipoim_niopalat 5.046 -0.050
## Subecolla_sitohcirt 5.045 -0.058
## Rumeloeahcra_irojam 5.054 -0.065
## Ihava_reginal 5.060 -0.057
## Ihava_silatnedicco 5.079 -0.046
## Suelagoriehc_iyelssorc 5.055 -0.061
## Suelagoriehc_rojam 5.057 -0.061
## Suelagoriehc_suidem 5.075 -0.059
## Ainotnebuad_sisneiracsagadam 5.068 -0.061
## Rumelue_retnevirbur 5.035 -0.058
## Rumelapah_sumis 5.031 -0.068
## Irdni_irdni 5.059 -0.049
## Rumelipel_silceea 5.050 -0.062
## Rumelipel_suniletsum 5.028 -0.087
## Rumelipel_ilosanairdnar 5.056 -0.135
## Rumelipel_sutaduacifur 5.034 -0.060
## Rumelipel_silanoirtnetpes 5.025 -0.083
## Subecorcim_eahtreb 5.042 -0.056
## Subecorcim_sisnevalognob 5.084 -0.073
## Subecorcim_sufuroesirg 5.058 -0.054
## Subecorcim_eaylloj 5.049 -0.057
## Subecorcim_sunixoym 5.052 -0.045
## Subecorcim_isnommis 5.035 -0.048
## Renahp_reficruf 5.016 -0.061
## Sucehtiporp_ilereuqoc 5.055 -0.042
## Sucehtiporp_illasrettat 5.069 -0.046
## Sucehtiporp_ixuaerrev 4.985 -0.070
## Aicerav_arbur 5.073 -0.068
## Attauola_ayarac 5.047 -0.070
## Attauola_ataillap 5.051 -0.062
## Seleta_specicsuf 5.001 -0.088
## Seleta_sucsinap 5.005 -0.076
## Seletyhcarb_sedionhcara 5.092 -0.015
## Xirhtogal_ahcirtogal 5.033 -0.091
## Sutoa_sutalufni 5.080 -0.061
## Sutoa_specirgin 5.067 -0.065
## Xirhtillac_eailime 5.015 -0.059
## Xirhtillac_atallicinep 5.049 -0.061
## Xirhtillac_aeamgyp 5.031 -0.063
## Subec_snorfibla 5.059 -0.058
## Suniugas_xatsym 5.097 -0.046
## Suniugas_supideo 5.027 -0.080
## Suniugas_sutitrapirt 5.027 -0.073
## Irimias_sisneivilob 5.062 -0.054
## Sirol_sunairekkedyl 5.051 -0.060
## Sirol_sudargidrat 5.051 -0.060
## Subecitcyn_sisnelagneb 5.069 -0.097
## Subecitcyn_gnacuoc 5.036 -0.037
## Subecitcyn_sucinavaj 5.080 -0.007
## Subecitcyn_sisneganem 5.041 -0.122
## Omoh_sneipas 5.129 -0.059
## Setabolyh_irelleum 5.059 -0.071
## Sucsamon_ealleirbag 5.073 -0.066
## Sucsamon_sutusan 5.045 -0.029
## Sucsamon_ikis 5.086 -0.053
## Ognop_sueamgyp 5.054 -0.062
## Aicehtip_aicehtip 5.047 -0.063
## Acacam_sediotcra 5.061 -0.066
## Acacam_sipolcyc 5.046 -0.081
## Acacam_siralucicsaf 5.048 -0.053
## Acacam_ikceh 5.035 -0.031
## Acacam_attalum 5.029 -0.056
## Acacam_anirtsemen 5.051 -0.046
## Acacam_snecsergin 5.068 -0.037
## Acacam_sunelis 5.095 -0.034
## Acacam_sunavlys 5.068 -0.060
## Acacam_anaeknot 5.037 -0.054
## Oipap_sibuna 5.047 -0.021
## Oipap_sulahpeconyc 5.071 -0.048
## Oipap_sayrdamah 5.040 -0.046
## Oipap_sunisru 5.052 -0.061
## Subecewgnur_ijnupik 5.063 -0.077
## Suboloc_sisnelogna 5.046 -0.062
## Suboloc_susorellev 5.048 -0.060
## Subolocoilip_muronodrog 5.063 -0.068
## Subolocoilip_issuerp 4.980 -0.019
## Subolocoilip_sutartimofur 4.951 0.019
## Subolocorp_surev 5.021 -0.069
## Xirhtagyp_aerenic 5.084 -0.070
## Sucehtiponihr_sulucnuva 5.005 -0.100
## Sucehtiponihr_ihcilerb 4.994 -0.104
## Sucehtipyhcart_iruocaled 5.031 -0.069
## Sucehtipyhcart_isiocnarf 5.040 -0.101
## Sucehtipyhcart_mutoal 5.044 -0.063
## Sucehtipyhcart_ieryahp 5.137 -0.058
## Sucitoue_sulutnagele 5.044 -0.073
## Ogalag_ieihcstam 5.049 -0.069
## Ogalag_sisnelagenes 4.986 -0.045
## Rumeloto_sutaduacissarc 5.042 -0.040
## Rumeloto_iittenrag 5.051 -0.065
## Suisrat_sunacnab 5.050 -0.062
## mating.systemuniM.uniF
## Sucehtipocrec_siralugobla 0.016
## Sucehtipocrec_suhpec 0.030
## Sucehtipocrec_itseohl 0.015
## Sucehtipocrec_anom 0.020
## Sucehtipocrec_atsiruatep 0.025
## Subecorolhc_surhtyregyp 0.013
## Subecorolhc_sulatnat -0.051
## Sucehtipoim_niopalat 0.024
## Subecolla_sitohcirt 0.030
## Rumeloeahcra_irojam 0.020
## Ihava_reginal 0.032
## Ihava_silatnedicco 0.045
## Suelagoriehc_iyelssorc 0.025
## Suelagoriehc_rojam 0.026
## Suelagoriehc_suidem 0.029
## Ainotnebuad_sisneiracsagadam 0.019
## Rumelue_retnevirbur 0.031
## Rumelapah_sumis 0.013
## Irdni_irdni 0.030
## Rumelipel_silceea 0.025
## Rumelipel_suniletsum 0.032
## Rumelipel_ilosanairdnar -0.040
## Rumelipel_sutaduacifur 0.038
## Rumelipel_silanoirtnetpes 0.018
## Subecorcim_eahtreb 0.024
## Subecorcim_sisnevalognob 0.017
## Subecorcim_sufuroesirg 0.006
## Subecorcim_eaylloj 0.027
## Subecorcim_sunixoym 0.023
## Subecorcim_isnommis 0.036
## Renahp_reficruf 0.034
## Sucehtiporp_ilereuqoc -0.001
## Sucehtiporp_illasrettat 0.005
## Sucehtiporp_ixuaerrev 0.045
## Aicerav_arbur 0.005
## Attauola_ayarac 0.033
## Attauola_ataillap 0.026
## Seleta_specicsuf 0.049
## Seleta_sucsinap 0.058
## Seletyhcarb_sedionhcara 0.055
## Xirhtogal_ahcirtogal 0.000
## Sutoa_sutalufni 0.025
## Sutoa_specirgin 0.023
## Xirhtillac_eailime 0.023
## Xirhtillac_atallicinep 0.027
## Xirhtillac_aeamgyp 0.023
## Subec_snorfibla 0.033
## Suniugas_xatsym 0.042
## Suniugas_supideo 0.009
## Suniugas_sutitrapirt 0.021
## Irimias_sisneivilob 0.035
## Sirol_sunairekkedyl 0.024
## Sirol_sudargidrat 0.022
## Subecitcyn_sisnelagneb 0.060
## Subecitcyn_gnacuoc -0.025
## Subecitcyn_sucinavaj 0.043
## Subecitcyn_sisneganem -0.033
## Omoh_sneipas 0.037
## Setabolyh_irelleum 0.025
## Sucsamon_ealleirbag 0.016
## Sucsamon_sutusan 0.069
## Sucsamon_ikis 0.041
## Ognop_sueamgyp 0.025
## Aicehtip_aicehtip 0.024
## Acacam_sediotcra 0.022
## Acacam_sipolcyc 0.006
## Acacam_siralucicsaf 0.023
## Acacam_ikceh 0.060
## Acacam_attalum -0.015
## Acacam_anirtsemen 0.031
## Acacam_snecsergin 0.054
## Acacam_sunelis 0.034
## Acacam_sunavlys 0.031
## Acacam_anaeknot 0.046
## Oipap_sibuna 0.074
## Oipap_sulahpeconyc 0.024
## Oipap_sayrdamah 0.060
## Oipap_sunisru 0.024
## Subecewgnur_ijnupik 0.030
## Suboloc_sisnelogna 0.027
## Suboloc_susorellev 0.028
## Subolocoilip_muronodrog 0.019
## Subolocoilip_issuerp 0.031
## Subolocoilip_sutartimofur 0.096
## Subolocorp_surev 0.004
## Xirhtagyp_aerenic 0.012
## Sucehtiponihr_sulucnuva -0.038
## Sucehtiponihr_ihcilerb -0.046
## Sucehtipyhcart_iruocaled 0.038
## Sucehtipyhcart_isiocnarf 0.085
## Sucehtipyhcart_mutoal 0.028
## Sucehtipyhcart_ieryahp 0.033
## Sucitoue_sulutnagele 0.020
## Ogalag_ieihcstam 0.021
## Ogalag_sisnelagenes 0.039
## Rumeloto_sutaduacissarc 0.040
## Rumeloto_iittenrag 0.022
## Suisrat_sunacnab 0.025
round(stability.ests$SEs, digits=3)#shows the standard errors obtained for each individual taxon exluded
## (Intercept) group.size longevity
## Sucehtipocrec_siralugobla 1.985 0.042 0.036
## Sucehtipocrec_suhpec 1.986 0.043 0.037
## Sucehtipocrec_itseohl 1.986 0.042 0.037
## Sucehtipocrec_anom 2.003 0.043 0.037
## Sucehtipocrec_atsiruatep 1.991 0.043 0.037
## Subecorolhc_surhtyregyp 1.992 0.043 0.037
## Subecorolhc_sulatnat 1.979 0.042 0.036
## Sucehtipoim_niopalat 1.974 0.042 0.036
## Subecolla_sitohcirt 1.993 0.043 0.037
## Rumeloeahcra_irojam 1.991 0.043 0.037
## Ihava_reginal 1.994 0.043 0.037
## Ihava_silatnedicco 1.987 0.042 0.037
## Suelagoriehc_iyelssorc 1.988 0.043 0.037
## Suelagoriehc_rojam 1.990 0.043 0.037
## Suelagoriehc_suidem 1.986 0.043 0.037
## Ainotnebuad_sisneiracsagadam 2.016 0.042 0.037
## Rumelue_retnevirbur 1.988 0.042 0.037
## Rumelapah_sumis 1.986 0.042 0.037
## Irdni_irdni 1.985 0.042 0.036
## Rumelipel_silceea 1.991 0.043 0.037
## Rumelipel_suniletsum 1.906 0.041 0.035
## Rumelipel_ilosanairdnar 1.962 0.042 0.036
## Rumelipel_sutaduacifur 1.996 0.043 0.037
## Rumelipel_silanoirtnetpes 1.953 0.042 0.036
## Subecorcim_eahtreb 1.994 0.043 0.037
## Subecorcim_sisnevalognob 1.984 0.042 0.036
## Subecorcim_sufuroesirg 1.987 0.043 0.036
## Subecorcim_eaylloj 1.992 0.043 0.037
## Subecorcim_sunixoym 1.968 0.042 0.036
## Subecorcim_isnommis 1.992 0.043 0.037
## Renahp_reficruf 1.979 0.042 0.036
## Sucehtiporp_ilereuqoc 2.001 0.043 0.037
## Sucehtiporp_illasrettat 2.005 0.043 0.037
## Sucehtiporp_ixuaerrev 1.994 0.043 0.037
## Aicerav_arbur 1.993 0.043 0.036
## Attauola_ayarac 1.987 0.042 0.037
## Attauola_ataillap 1.992 0.043 0.037
## Seleta_specicsuf 1.974 0.042 0.036
## Seleta_sucsinap 1.994 0.043 0.037
## Seletyhcarb_sedionhcara 1.984 0.042 0.037
## Xirhtogal_ahcirtogal 2.006 0.043 0.037
## Sutoa_sutalufni 1.984 0.042 0.037
## Sutoa_specirgin 1.994 0.042 0.038
## Xirhtillac_eailime 2.007 0.043 0.037
## Xirhtillac_atallicinep 2.012 0.043 0.037
## Xirhtillac_aeamgyp 1.981 0.042 0.036
## Subec_snorfibla 2.002 0.044 0.037
## Suniugas_xatsym 1.994 0.043 0.037
## Suniugas_supideo 2.005 0.044 0.037
## Suniugas_sutitrapirt 1.993 0.043 0.037
## Irimias_sisneivilob 1.992 0.043 0.037
## Sirol_sunairekkedyl 1.991 0.042 0.037
## Sirol_sudargidrat 1.989 0.042 0.037
## Subecitcyn_sisnelagneb 1.986 0.042 0.037
## Subecitcyn_gnacuoc 1.990 0.042 0.037
## Subecitcyn_sucinavaj 1.954 0.042 0.036
## Subecitcyn_sisneganem 1.977 0.042 0.036
## Omoh_sneipas 1.973 0.042 0.037
## Setabolyh_irelleum 1.991 0.043 0.037
## Sucsamon_ealleirbag 1.988 0.043 0.037
## Sucsamon_sutusan 1.986 0.043 0.036
## Sucsamon_ikis 1.978 0.042 0.036
## Ognop_sueamgyp 1.996 0.043 0.037
## Aicehtip_aicehtip 2.003 0.043 0.037
## Acacam_sediotcra 1.996 0.043 0.037
## Acacam_sipolcyc 1.986 0.042 0.037
## Acacam_siralucicsaf 1.987 0.042 0.037
## Acacam_ikceh 1.980 0.042 0.036
## Acacam_attalum 1.949 0.042 0.036
## Acacam_anirtsemen 1.984 0.042 0.036
## Acacam_snecsergin 1.990 0.043 0.037
## Acacam_sunelis 1.973 0.042 0.036
## Acacam_sunavlys 1.989 0.043 0.037
## Acacam_anaeknot 1.990 0.043 0.037
## Oipap_sibuna 2.002 0.043 0.037
## Oipap_sulahpeconyc 1.990 0.042 0.037
## Oipap_sayrdamah 1.997 0.043 0.037
## Oipap_sunisru 1.993 0.043 0.037
## Subecewgnur_ijnupik 1.985 0.042 0.037
## Suboloc_sisnelogna 2.010 0.043 0.037
## Suboloc_susorellev 1.981 0.042 0.036
## Subolocoilip_muronodrog 2.002 0.043 0.037
## Subolocoilip_issuerp 1.947 0.043 0.036
## Subolocoilip_sutartimofur 1.966 0.042 0.036
## Subolocorp_surev 1.952 0.042 0.036
## Xirhtagyp_aerenic 1.992 0.042 0.037
## Sucehtiponihr_sulucnuva 1.980 0.043 0.036
## Sucehtiponihr_ihcilerb 1.972 0.042 0.036
## Sucehtipyhcart_iruocaled 1.986 0.042 0.037
## Sucehtipyhcart_isiocnarf 1.955 0.042 0.037
## Sucehtipyhcart_mutoal 1.998 0.043 0.037
## Sucehtipyhcart_ieryahp 1.981 0.042 0.037
## Sucitoue_sulutnagele 2.063 0.044 0.039
## Ogalag_ieihcstam 2.046 0.044 0.038
## Ogalag_sisnelagenes 1.982 0.042 0.036
## Rumeloto_sutaduacissarc 1.995 0.042 0.037
## Rumeloto_iittenrag 1.993 0.043 0.037
## Suisrat_sunacnab 2.037 0.043 0.037
## log.body.size mating.systemmultiF.uniM
## Sucehtipocrec_siralugobla 0.302 0.203
## Sucehtipocrec_suhpec 0.299 0.203
## Sucehtipocrec_itseohl 0.299 0.203
## Sucehtipocrec_anom 0.299 0.203
## Sucehtipocrec_atsiruatep 0.300 0.203
## Subecorolhc_surhtyregyp 0.308 0.207
## Subecorolhc_sulatnat 0.301 0.207
## Sucehtipoim_niopalat 0.297 0.202
## Subecolla_sitohcirt 0.301 0.204
## Rumeloeahcra_irojam 0.300 0.204
## Ihava_reginal 0.302 0.204
## Ihava_silatnedicco 0.303 0.204
## Suelagoriehc_iyelssorc 0.299 0.203
## Suelagoriehc_rojam 0.301 0.203
## Suelagoriehc_suidem 0.301 0.203
## Ainotnebuad_sisneiracsagadam 0.299 0.203
## Rumelue_retnevirbur 0.301 0.203
## Rumelapah_sumis 0.299 0.202
## Irdni_irdni 0.299 0.203
## Rumelipel_silceea 0.301 0.206
## Rumelipel_suniletsum 0.287 0.195
## Rumelipel_ilosanairdnar 0.295 0.204
## Rumelipel_sutaduacifur 0.301 0.203
## Rumelipel_silanoirtnetpes 0.293 0.199
## Subecorcim_eahtreb 0.300 0.203
## Subecorcim_sisnevalognob 0.301 0.203
## Subecorcim_sufuroesirg 0.298 0.202
## Subecorcim_eaylloj 0.300 0.204
## Subecorcim_sunixoym 0.296 0.201
## Subecorcim_isnommis 0.303 0.207
## Renahp_reficruf 0.299 0.202
## Sucehtiporp_ilereuqoc 0.299 0.204
## Sucehtiporp_illasrettat 0.302 0.206
## Sucehtiporp_ixuaerrev 0.303 0.202
## Aicerav_arbur 0.300 0.203
## Attauola_ayarac 0.299 0.204
## Attauola_ataillap 0.300 0.203
## Seleta_specicsuf 0.299 0.202
## Seleta_sucsinap 0.301 0.203
## Seletyhcarb_sedionhcara 0.301 0.208
## Xirhtogal_ahcirtogal 0.301 0.210
## Sutoa_sutalufni 0.299 0.202
## Sutoa_specirgin 0.299 0.202
## Xirhtillac_eailime 0.301 0.202
## Xirhtillac_atallicinep 0.300 0.203
## Xirhtillac_aeamgyp 0.299 0.202
## Subec_snorfibla 0.300 0.203
## Suniugas_xatsym 0.301 0.203
## Suniugas_supideo 0.301 0.205
## Suniugas_sutitrapirt 0.299 0.203
## Irimias_sisneivilob 0.300 0.203
## Sirol_sunairekkedyl 0.299 0.203
## Sirol_sudargidrat 0.298 0.202
## Subecitcyn_sisnelagneb 0.300 0.208
## Subecitcyn_gnacuoc 0.300 0.206
## Subecitcyn_sucinavaj 0.294 0.202
## Subecitcyn_sisneganem 0.298 0.209
## Omoh_sneipas 0.300 0.201
## Setabolyh_irelleum 0.300 0.204
## Sucsamon_ealleirbag 0.301 0.203
## Sucsamon_sutusan 0.299 0.206
## Sucsamon_ikis 0.299 0.202
## Ognop_sueamgyp 0.302 0.203
## Aicehtip_aicehtip 0.301 0.204
## Acacam_sediotcra 0.302 0.204
## Acacam_sipolcyc 0.299 0.205
## Acacam_siralucicsaf 0.299 0.204
## Acacam_ikceh 0.299 0.205
## Acacam_attalum 0.294 0.199
## Acacam_anirtsemen 0.299 0.204
## Acacam_snecsergin 0.299 0.205
## Acacam_sunelis 0.299 0.203
## Acacam_sunavlys 0.303 0.203
## Acacam_anaeknot 0.301 0.204
## Oipap_sibuna 0.299 0.213
## Oipap_sulahpeconyc 0.304 0.206
## Oipap_sayrdamah 0.300 0.205
## Oipap_sunisru 0.303 0.203
## Subecewgnur_ijnupik 0.299 0.203
## Suboloc_sisnelogna 0.301 0.203
## Suboloc_susorellev 0.298 0.202
## Subolocoilip_muronodrog 0.306 0.206
## Subolocoilip_issuerp 0.293 0.199
## Subolocoilip_sutartimofur 0.302 0.206
## Subolocorp_surev 0.293 0.198
## Xirhtagyp_aerenic 0.306 0.204
## Sucehtiponihr_sulucnuva 0.300 0.204
## Sucehtiponihr_ihcilerb 0.300 0.204
## Sucehtipyhcart_iruocaled 0.300 0.203
## Sucehtipyhcart_isiocnarf 0.294 0.201
## Sucehtipyhcart_mutoal 0.302 0.203
## Sucehtipyhcart_ieryahp 0.302 0.201
## Sucitoue_sulutnagele 0.301 0.209
## Ogalag_ieihcstam 0.300 0.212
## Ogalag_sisnelagenes 0.306 0.203
## Rumeloto_sutaduacissarc 0.302 0.224
## Rumeloto_iittenrag 0.300 0.223
## Suisrat_sunacnab 0.301 0.203
## mating.systemuniM.uniF
## Sucehtipocrec_siralugobla 0.217
## Sucehtipocrec_suhpec 0.217
## Sucehtipocrec_itseohl 0.217
## Sucehtipocrec_anom 0.217
## Sucehtipocrec_atsiruatep 0.217
## Subecorolhc_surhtyregyp 0.226
## Subecorolhc_sulatnat 0.226
## Sucehtipoim_niopalat 0.215
## Subecolla_sitohcirt 0.218
## Rumeloeahcra_irojam 0.218
## Ihava_reginal 0.219
## Ihava_silatnedicco 0.219
## Suelagoriehc_iyelssorc 0.217
## Suelagoriehc_rojam 0.217
## Suelagoriehc_suidem 0.217
## Ainotnebuad_sisneiracsagadam 0.216
## Rumelue_retnevirbur 0.217
## Rumelapah_sumis 0.216
## Irdni_irdni 0.217
## Rumelipel_silceea 0.217
## Rumelipel_suniletsum 0.208
## Rumelipel_ilosanairdnar 0.217
## Rumelipel_sutaduacifur 0.219
## Rumelipel_silanoirtnetpes 0.212
## Subecorcim_eahtreb 0.217
## Subecorcim_sisnevalognob 0.217
## Subecorcim_sufuroesirg 0.217
## Subecorcim_eaylloj 0.217
## Subecorcim_sunixoym 0.215
## Subecorcim_isnommis 0.219
## Renahp_reficruf 0.216
## Sucehtiporp_ilereuqoc 0.219
## Sucehtiporp_illasrettat 0.221
## Sucehtiporp_ixuaerrev 0.216
## Aicerav_arbur 0.218
## Attauola_ayarac 0.217
## Attauola_ataillap 0.218
## Seleta_specicsuf 0.216
## Seleta_sucsinap 0.218
## Seletyhcarb_sedionhcara 0.218
## Xirhtogal_ahcirtogal 0.221
## Sutoa_sutalufni 0.216
## Sutoa_specirgin 0.216
## Xirhtillac_eailime 0.216
## Xirhtillac_atallicinep 0.217
## Xirhtillac_aeamgyp 0.216
## Subec_snorfibla 0.218
## Suniugas_xatsym 0.216
## Suniugas_supideo 0.218
## Suniugas_sutitrapirt 0.216
## Irimias_sisneivilob 0.217
## Sirol_sunairekkedyl 0.217
## Sirol_sudargidrat 0.216
## Subecitcyn_sisnelagneb 0.222
## Subecitcyn_gnacuoc 0.228
## Subecitcyn_sucinavaj 0.213
## Subecitcyn_sisneganem 0.222
## Omoh_sneipas 0.215
## Setabolyh_irelleum 0.217
## Sucsamon_ealleirbag 0.217
## Sucsamon_sutusan 0.222
## Sucsamon_ikis 0.216
## Ognop_sueamgyp 0.217
## Aicehtip_aicehtip 0.217
## Acacam_sediotcra 0.217
## Acacam_sipolcyc 0.219
## Acacam_siralucicsaf 0.217
## Acacam_ikceh 0.219
## Acacam_attalum 0.214
## Acacam_anirtsemen 0.217
## Acacam_snecsergin 0.219
## Acacam_sunelis 0.215
## Acacam_sunavlys 0.218
## Acacam_anaeknot 0.223
## Oipap_sibuna 0.231
## Oipap_sulahpeconyc 0.217
## Oipap_sayrdamah 0.225
## Oipap_sunisru 0.220
## Subecewgnur_ijnupik 0.216
## Suboloc_sisnelogna 0.217
## Suboloc_susorellev 0.216
## Subolocoilip_muronodrog 0.219
## Subolocoilip_issuerp 0.211
## Subolocoilip_sutartimofur 0.218
## Subolocorp_surev 0.212
## Xirhtagyp_aerenic 0.218
## Sucehtiponihr_sulucnuva 0.222
## Sucehtiponihr_ihcilerb 0.221
## Sucehtipyhcart_iruocaled 0.217
## Sucehtipyhcart_isiocnarf 0.215
## Sucehtipyhcart_mutoal 0.218
## Sucehtipyhcart_ieryahp 0.215
## Sucitoue_sulutnagele 0.218
## Ogalag_ieihcstam 0.219
## Ogalag_sisnelagenes 0.217
## Rumeloto_sutaduacissarc 0.227
## Rumeloto_iittenrag 0.227
## Suisrat_sunacnab 0.217
#note that in these latter two outputs the rows for species where the function pgls failed to converge would comprise NAs
Full null model comparison to establish the significance of the test predictors as a whole:
#run null model comprising only the control predictor 'body size':
null=pgls(brain.size~log.body.size, data=test.data, lambda="ML")
#compare null and full model using a an F-test:
anova(null, full, test="F")
## Analysis of Variance Table
## pgls: lambda = 1.00, delta = 1.00, kappa = 1.00
##
## Model 1: brain.size ~ log.body.size
## Model 2: brain.size ~ group.size + longevity + log.body.size + mating.system
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 96 45.8
## 2 92 12.6 4 33.2 60.5 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#is clearly significant; the test predictors as a whole have an impact on brain size
Inference about individual terms:
summary(full)$coefficients
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 6.41428 1.98015 3.2393 0.001668
## group.size 0.59996 0.04228 14.1916 0.000000
## longevity -0.02835 0.03640 -0.7789 0.438050
## log.body.size 5.05064 0.29809 16.9432 0.000000
## mating.systemmultiF.uniM -0.06138 0.20222 -0.3035 0.762185
## mating.systemuniM.uniF 0.02497 0.21600 0.1156 0.908205
#reveals the test predictor group size to have a positive and significant impact on brain size,
#and longevity to be non-significant
#furthermore, as expected the control predictor body size has a clearly significant positive impact on brain size.
#The predictor mating system is represented by two dummy variables; to get an overall p-value for this factor
#first run a model lacking mating system but comprising all other terms present in the full model:
red=pgls(brain.size ~ group.size + longevity + log.body.size, data=test.data, lambda="ML")
#then compare it with the full model using an F-test:
anova(red, full, test="F")
## Analysis of Variance Table
## pgls: lambda = 1.00, delta = 1.00, kappa = 1.00
##
## Model 1: brain.size ~ group.size + longevity + log.body.size
## Model 2: brain.size ~ group.size + longevity + log.body.size + mating.system
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 94 12.7
## 2 92 12.6 2 0.0349 0.13 0.88
#reveals that mating system has no significant impact on the response
Dealing further with colliearity
Above it appeared that the test predictor home range size and the control predictor longevity were pretty collinear. To explore this issue a little further, one could fit an additional model including both predictors (and also all other terms present in the original model) and also a model in which home range size replaces body size.
#fit a model with both body size and home range size included:
full.with.hr.bo=pgls(brain.size ~ group.size + longevity + log.body.size + mating.system + hr.size, data=test.data, lambda="ML")
#inspect the derived estimates:
round(summary(full.with.hr.bo)$coefficients, digits=3)
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 5.238 2.280 2.297 0.024
## group.size 0.606 0.043 14.213 0.000
## longevity -0.026 0.036 -0.703 0.484
## log.body.size 4.644 0.492 9.440 0.000
## mating.systemmultiF.uniM -0.062 0.202 -0.307 0.760
## mating.systemuniM.uniF 0.007 0.217 0.032 0.975
## hr.size 0.030 0.029 1.039 0.302
#and compare them with those obtained for the original full model:
round(summary(full)$coefficients, digits=3)
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 6.414 1.980 3.239 0.002
## group.size 0.600 0.042 14.192 0.000
## longevity -0.028 0.036 -0.779 0.438
## log.body.size 5.051 0.298 16.943 0.000
## mating.systemmultiF.uniM -0.061 0.202 -0.304 0.762
## mating.systemuniM.uniF 0.025 0.216 0.116 0.908
#the effect of body size did not change much, and home range size does not seem to have an obvious impact
#fit an additional model including home range but not body size:
full.with.hr=pgls(brain.size ~ group.size + longevity + mating.system + hr.size, data=test.data, lambda="ML")
#inspect the derived estimates:
round(summary(full.with.hr)$coefficients, digits=3)
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -3.171 2.819 -1.125 0.264
## group.size 0.642 0.058 10.999 0.000
## longevity 0.005 0.051 0.097 0.923
## mating.systemmultiF.uniM -0.160 0.316 -0.507 0.613
## mating.systemuniM.uniF -0.103 0.337 -0.306 0.760
## hr.size 0.246 0.024 10.360 0.000
#this latter model reveals home range to have a clear (i.e., significant) positive impact on brain size
From this last section of code I would conclude that it might well be that home range size and brain size are indeed correlated, but the current data do not allow to reliably tease apart the effects of home range size and body size on brain size.
References
- Arnold C, Matthews LJ & Nunn CL. 2010. The 10kTrees Website: A New Online Resource for Primate Phylogeny. Evolutionary Anthropology 19, 114-118.
- Fox J & Weisberg S. 2011. An R Companion to Applied Regression. Second edition. Sage, Thousand Oaks, CA.
- Orme D, Freckleton R, Thomas G, Petzoldt T, Fritz S, Isaac N & Pearse W. 2013. caper: Comparative Analyses of Phylogenetics and Evolution in R. R package version 0.5.2.
- Paradis E, Claude J & Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289-290.