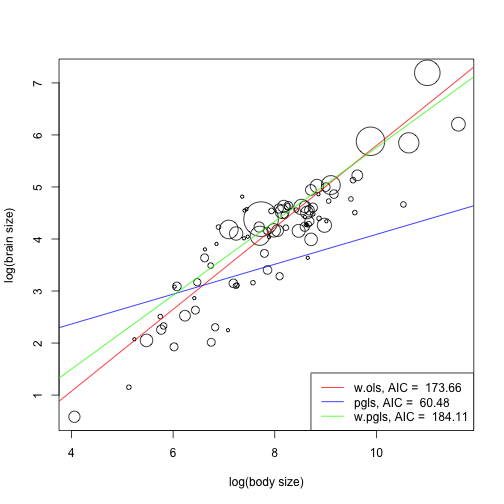
plot(data.spec$lg.body, data.spec$lg.brain, pch = 1, cex = 0.6 * sqrt(data.spec$brain_N), xlab = "log(body size)", ylab = "log(brain size)") abline(w.ols, col = "red") abline(pgls, col = "blue") abline(w.pgls, col = "green") legend(x = "bottomright", legend = c(paste("w.ols, AIC = ", round(AIC(w.ols), 2)), paste("pgls, AIC = ", round(AIC(pgls), 2)), paste("w.pgls, AIC = ", round(AIC(w.pgls), 2))), lty = 1, col = c("red", "blue", "green"))