Sources
R packages
"ape" (Paradis et al 2004)
"phytools" (Revell 2012)
Data
brain size and body size on primates ("primate_ind.txt", a tab separated text file), with more than one observation for the majority of species, individual-specific data
brain size and body size on primates ("primate_spec.txt", a tab separated text file), species-specific data obtained from the exercises above, includes information on within-species sampling (sample sizes, variances and standard errors
phylogeny ("primate_tree.phy", in phylip format) is taken from 10kTrees (Arnold et al 2010) and tailored to the data at hand
Codes
Estimating species-specific means and variances, ancestral states and evolutionary rates by using Bayesian MCMC
Model assuming common variance
data.ind = read.table("primate_ind.txt", sep = "\t", header = TRUE)
library(ape)
tree = read.tree("primate_tree.phy")
brain = c(data.ind$brain)
names(brain) = data.ind$species
library(phytools, quietly = T)
fittedB = fitBayes(tree, brain, ngen = 10000, method = "reduced")
## Control parameters (set by user or default):
## List of 8
## $ sig2 : num 1249
## $ a : num 93.7
## $ xbar : Named num [1:86] 57 55.7 69.1 64.1 57.5 ...
## ..- attr(*, "names")= chr [1:86] "Allenopithecus_nigroviridis" "Cercopithecus_albogularis" "Cercopithecus_mitis" "Cercopithecus_ascanius" ...
## $ intV : num 455
## $ pr.mean: num [1:89] 1000 0 0 0 0 0 0 0 0 0 ...
## $ pr.var : num [1:89] 1e+06 1e+03 1e+03 1e+03 1e+03 1e+03 1e+03 1e+03 1e+03 1e+03 ...
## $ prop : num [1:89] 12.5 12.5 912 912 912 ...
## $ sample : num 100
## Starting MCMC...
## Done MCMC.
est.means = colMeans(fittedB[21:101, c(-1, -2, -3, -(dim(fittedB)[2] - 1), -dim(fittedB)[2])])
art.means = tapply(data.ind$brain, data.ind$species, mean)
plot(art.means, est.means[names(art.means)], xlab = "Arithmetic means", ylab = "Bayesian estimates of means")
abline(a = 0, b = 1, col = "red")
legend(x = "bottomright", legend = c(paste("var = ", round(mean(fittedB[21:101,
(dim(fittedB)[2] - 1)]), 2)), paste("MCRA = ", round(mean(fittedB[21:101,
3]), 2)), paste("sigma^2 = ", round(mean(fittedB[21:101, 2]), 2))))
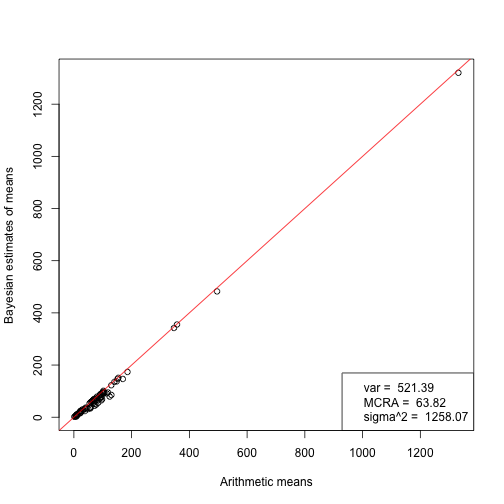
The cicles are species-specific means as can be calculated from the raw individual-specific data by using their arithemtic means or Bayesian MCMC-based estimates that incorporate phylogenetic constraints. Legend on the bottom right shows the assumed common intraspecific variance, trait value at the root of the phylogeny and evolutionary rate.
Model assuming heterogeneous variance
fittedB = fitBayes(tree, brain, ngen = 10000, method = "full")
## Control parameters (set by user or default):
## List of 8
## $ sig2 : num 1249
## $ a : num 93.7
## $ xbar : Named num [1:86] 57 55.7 69.1 64.1 57.5 ...
## ..- attr(*, "names")= chr [1:86] "Allenopithecus_nigroviridis" "Cercopithecus_albogularis" "Cercopithecus_mitis" "Cercopithecus_ascanius" ...
## $ v : Named num [1:86] 455 455 455 455 455 ...
## ..- attr(*, "names")= chr [1:86] "Allenopithecus_nigroviridis" "Cercopithecus_albogularis" "Cercopithecus_mitis" "Cercopithecus_ascanius" ...
## $ pr.mean: num [1:174] 1000 0 0 0 0 0 0 0 0 0 ...
## $ pr.var : num [1:174] 1e+06 1e+03 1e+03 1e+03 1e+03 1e+03 1e+03 1e+03 1e+03 1e+03 ...
## $ prop : Named num [1:174] 12.5 12.5 912 912 912 ...
## ..- attr(*, "names")= chr [1:174] "" "" "" "" ...
## $ sample : num 100
## Starting MCMC...
## Done MCMC.
est.means = colMeans(fittedB[21:101, 4:((dim(fittedB)[2] - 4)/2 + 3)])
plot(art.means, est.means[names(art.means)], xlab = "Arithmetic means", ylab = "Bayesian estimates of means")
abline(a = 0, b = 1, col = "red")
legend(x = "bottomright", legend = c(paste("MCRA = ", round(mean(fittedB[21:101,
3]), 2)), paste("sigma^2 = ", round(mean(fittedB[21:101, 2]), 2))))
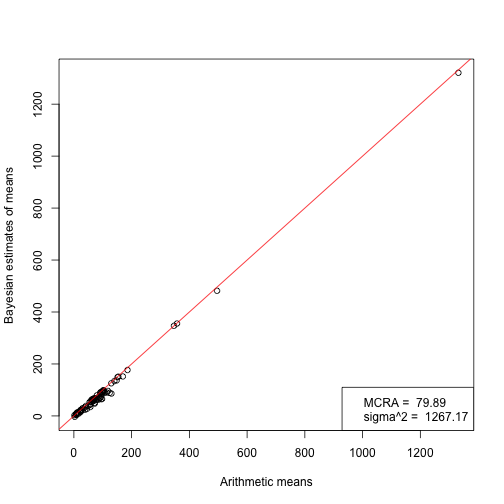
The cicles are species-specific means as can be calculated from the raw individual-specific data by using their arithemtic means or Bayesian MCMC-based estimates that incorporate phylogenetic constraints. Legend on the bottom right shows trait value at the root of the phylogeny and evolutionary rate. Within-species variance is assumed to be different across species.
Estimating phylogenetic signal
Without implementing measurement errors
Blomberg's K
data.spec = read.table("primate_spec.txt", sep = "\t", header = TRUE)
library(ape)
tree = read.tree("primate_tree.phy")
phylosig(tree, brain, test = T)
## $K
## [1] 0.3145
##
## $P
## [1] 0.045
Pagel's lambda
phylosig(tree, brain, method = "lambda", test = T)
## $lambda
## [1] 1.012
##
## $logL
## [1] -534.4
##
## $logL0
## [1] -553.3
##
## $P
## [1] 7.47e-10
By implementing measurement errors
Blomberg' K
brain_se = c(data.spec$brain_se.adj)
names(brain_se) = data.spec$species
phylosig(tree, brain, test = T, se = brain_se)
## $K
## [1] 0.3237
##
## $P
## [1] 0.045
##
## $sig2
## [1] 1331
##
## $logL
## [1] -536.6
Pagel's lambda
phylosig(tree, brain, method = "lambda", test = T, se = brain_se)
## $lambda
## [1] 1.012
##
## $sig2
## [1] 1571
##
## $logL
## [1] -531.9
##
## $convergence
## [1] 0
##
## $message
## [1] "CONVERGENCE: REL_REDUCTION_OF_F <= FACTR*EPSMCH"
##
## $logL0
## [1] -552.7
##
## $P
## [1] 1.037e-10
References
- Arnold C, Matthews LJ, Nunn CL (2010) The 10kTrees website: a new online resource for primate hylogeny. Evol Anthropol 19: 114-118.
- Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289-290.
- Revell LJ (2012) Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3: 217-223.

